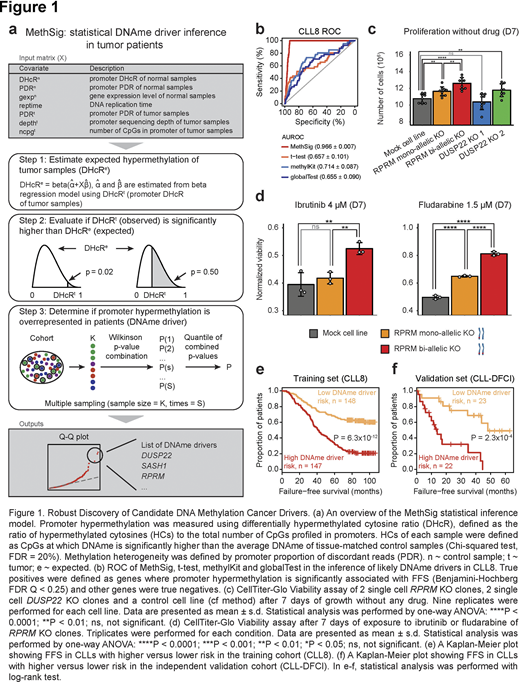
The field of cancer genomics has been empowered by increasingly sophisticated inference tools to distinguish driver mutations from the vastly greater number of passenger mutations. Epigenetic alterations such as promoter DNA hypermethylation have been shown to drive cancer through inactivation of tumor suppressor genes (TSGs), but growing malignant populations also accrue pervasive stochastic epigenetic changes in DNA methylation (DNAme), most of which likely carry little functional impact. Unlike with somatic mutations, we have limited ability to robustly differentiate driver DNAme changes (DNAme drivers) from stochastic, passenger DNAme changes. To address this challenge, we developed MethSig, a statistical inference framework that accounts for the varying stochastic hypermethylation rates across the genome and between samples. MethSig estimates expected background DNAme changes, thereby allowing the identification of epigenetically disrupted loci, where observed hypermethylation significantly exceeds expectation, potentially reflecting positive selection (Fig. 1a).
We applied MethSig to reduced representation bisulfite sequencing (RRBS) data of chronic lymphocytic leukemia (CLL) cohorts, which include 304 CLLs collected in a prospective clinical trial (CLL8) and 103 CLLs in a previously published study (CLL-DFCI, Landau et al., 2014), as well as other malignancies where RRBS data is available, including ductal carcinoma in situ (Abba et al., 2015) and multiple myeloma. Area under the receiver operating characteristic curve (AUROC) was used to evaluate sensitivity and specificity of methods in the inference of likely DNAme drivers. We identified two key features that are likely to be strongly associated with true candidate DNAme drivers: gene silencing in relation to promoter hypermethylation and association with clinical outcome. MethSig qualitatively improved ROC across those clinical and biological read outs (0.955 of MethSig, 95% confidence interval [CI] 0.945 - 0.965, versus 0.703 of benchmarked methods, 95% CI 0.669 - 0.737, Fig. 1b used CLL8 as an example).
We identified 189 candidate DNAme drivers in CLL, which include known TSGs, and are enriched in genes hypermethylated or inactivated across cancer types. To further validate MethSig's inferences, selected CLL candidate DNAme drivers (DUSP22, RPRM) underwent CRISPR/Cas9 knockout (KO) in CLL cells and stable KO clones were generated through single-cell cloning to eliminate genetic heterogeneity effect. The RPRM and DUSP22 KO clones showed faster growth without treatment (Fig. 1c) and superior fitness in ibrutinib/fludarabine treatment compared with controls (Fig. 1d). Notably, we observed a gene dose effect in the RPRM KO clones (Fig. 1c-d, greater growth of the bi-allelic compared to mono-allelic KO).
Elastic net regression with a Cox proportional hazards model was used to evaluate DNAme drivers' contribution to the prediction of failure-free survival after treatment (FFS; failure defined as retreatment or death) and a rigorous training (CLL8) and validation (an independent cohort CLL-DFCI) cohort study design was implemented to safeguard from overfitting and poor generalizability. DNAme drivers were found to be associated with shorter FFS in independent CLL cohorts (Fig. 1e-f). A regression model including established CLL risk indicators (IGHV unmutated status, del[17p] or TP53 mutation) showed an adjusted hazard ratio of 2.3 (95% CI 1.6 - 3.3, P = 2 × 10-6) in CLL8 cohort and 3.2 (95% CI 1.2 - 8.8, P = 0.02) in CLL-DFCI cohort for patients with high risk. Application of MethSig to CLL relapsed after chemoimmunotherapy further identified relapse-specific DNAme drivers, enriched in TP53 targets as well as DNA damage pathway, which indicates that CLL relapse after chemotherapy may follow an alternative path compared to CLL progression in the absence of therapy, offering novel insights for therapeutic strategies to address drug-resistant or relapsed cancer.
Collectively, our data support a novel framework for the analysis of DNAme changes in cancer to specifically identify DNAme drivers of disease progression and relapse, empowering the discovery of epigenetic mechanisms that enhance cancer cell fitness. This work addressed a central gap between cancer epigenetics and cancer genetics, where such tools have had a transformative impact in precision oncology and cancer gene discovery.
Tausch:Janssen-Cilag: Consultancy, Honoraria, Research Funding; Roche: Consultancy, Honoraria, Research Funding; AbbVie: Consultancy, Honoraria, Research Funding. Fink:AbbVie: Other: travel grants; Janssen: Honoraria; Celgene: Research Funding. Fischer:AbbVie: Honoraria; F. Hoffmann-La Roche: Honoraria, Other: travel grants. Gnirke:FL67 Inc.: Consultancy. Moreaux:Diag2Tec: Consultancy. Hallek:Celgene: Consultancy, Research Funding; Pharmacyclics: Consultancy, Research Funding; AbbVie: Consultancy, Research Funding; Gilead: Consultancy, Research Funding; Mundipharma: Consultancy, Research Funding; Janssen: Consultancy, Research Funding; Roche: Consultancy, Research Funding. Stilgenbauer:Janssen-Cilag: Consultancy, Honoraria, Other: travel support, Research Funding; Genentech: Consultancy, Honoraria, Other: travel support, Research Funding; Gilead: Consultancy, Honoraria, Other: travel support, Research Funding; Genzyme: Consultancy, Honoraria, Other: travel support, Research Funding; Pharmacyclics: Consultancy, Honoraria, Other, Research Funding; F. Hoffmann-LaRoche: Consultancy, Honoraria, Other: travel support, Research Funding; Celgene: Consultancy, Honoraria, Other: travel support, Research Funding; Boehringer-Ingelheim: Consultancy, Honoraria, Other: travel support, Research Funding; Amgen: Consultancy, Honoraria, Other: travel support, Research Funding; AbbVie: Consultancy, Honoraria, Other: travel support, Research Funding; GlaxoSmithKline: Consultancy, Honoraria, Other: travel support, Research Funding; Novartis: Consultancy, Honoraria, Other, Research Funding; Mundipharma: Consultancy, Honoraria, Other, Research Funding. Wu:BionTech: Current equity holder in publicly-traded company; Pharmacyclics: Research Funding. Elemento:Acuamark: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; OneThree Biotech: Current equity holder in private company, Other: Cofounder; Owkin: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; Sanofi: Research Funding; Eli Lilly: Research Funding; Volastra Therapeutics: Current equity holder in private company, Other: Cofounder; Freenome: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; Genetic Intelligence: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; Janssen: Research Funding. Landau:Bristol Myers Squibb: Research Funding; Illumina: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.